Computational Graph
SINGA can buffering operations to create a computational graph (CG). With the computational graph, SINGA can schedule the execution of operations as well as the memory allocation and release. It makes training more efficient while using less memory.
About Computational Graph
Introduction
Computational graph is used to represent networks of the flow of computation. It is composed of many nodes and edges, where nodes represent various operations and edges represent data. In deep neural networks, nodes are tensor-based operations such as convolution and edges are tensors.
The entire neural network is equivalent to a computational graph, all neural networks can correspond to a calculation graph. By representing the neural network as a calculation graph, some optimizations for neural networks can be performed on the calculation graph.
Pipeline
The whole process of using the calculational graph to represent the model and execute the graph consists of roughly four steps. The whole process is actually similar to compiling. We first describe the program with code, then translate the program into intermediate code, then optimize the intermediate code and finally come up with many ways to efficiently execute the code. In neural networks, the intermediate code is the calculation graph. We can optimize through techniques like common sub-expression elimination. When the computer executes the compiled binary file, it can be efficiently executed by using multi-thread technology, and the same as the execution of the calculation graph. Therefore, some ideas of compilation principles can also be used in the optimization of calculation graphs.
Write the python code for the model.
Construct the computational graph based on the python code.
Optimize the computational graph.
Execute the computational graph efficiently.
Figure 1 shows a simple example of going through the entire process.

Figure 1 - The pipeline of using computational graph
An example of MLP
A simple MLP model can be constructed on the Python side by using some APIs of SINGA.
x = autograd.matmul(inputs, w0)
x = autograd.add_bias(x, b0)
x = autograd.relu(x)
x = autograd.matmul(x, w1)
x = autograd.add_bias(x, b1)
loss = autograd.softmax_cross_entropy(x, target)
sgd.backward_and_update(loss)
When the model is defined, there is actually a calculation graph corresponding to it. This calculation graph contains the calculations that the entire SINGA will perform. Figure 2 shows the computational graph corresponding to the MLP model defined above.
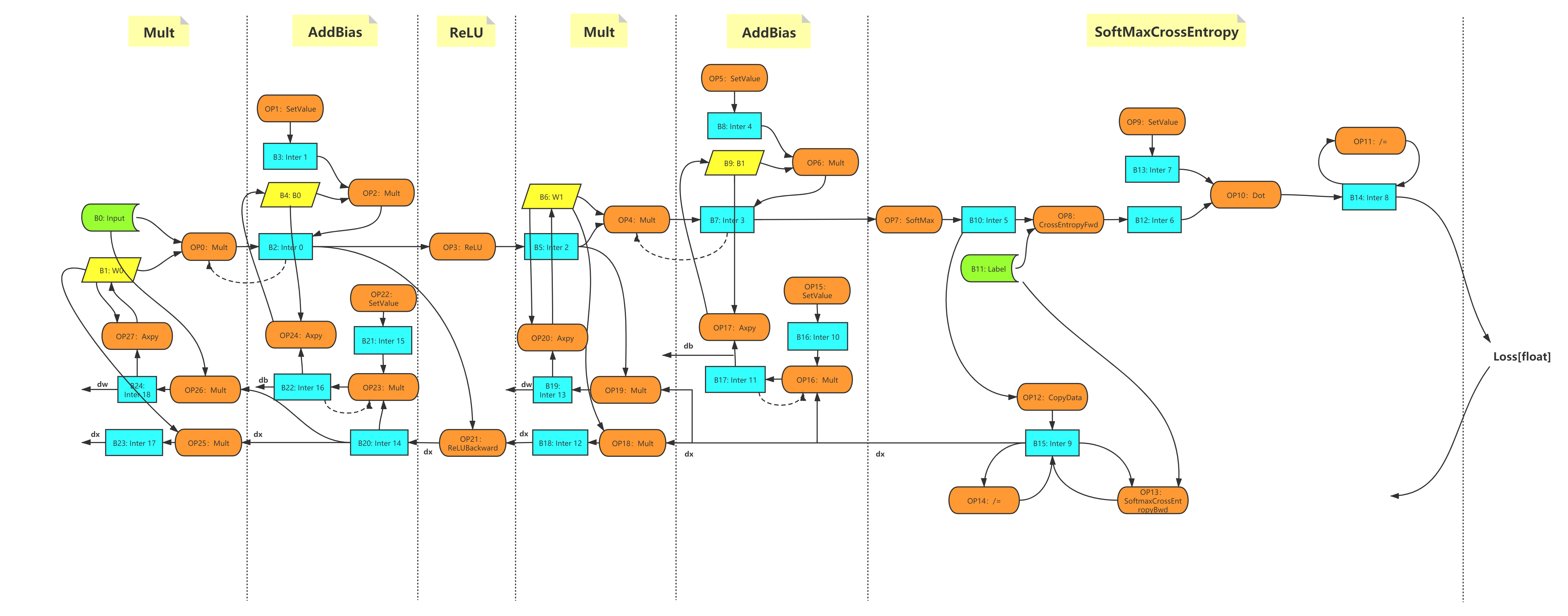
Figure 2 - The computational graph of MLP
Features
There are four main components of a computational graph in SINGA, namely (i) Computational graph construction, (ii) Lazy allocation, (iii) Automatic recycling, (iv) Shared memory. Details are as follows:
Computational graph construction: Construct a computational graph based on the mathematical or deep learning operations, and then run the graph to accomplish the training task. The computational graph also includes operations like communicator.synch and communicator.fusedSynch for the distributed training.Lazy allocation: When blocks are allocated, devices do not allocate memory for them immediately. Devices do memory allocation only when an operation uses this block for the first time.Automatic recycling: When we are running a graph in an iteration, it automatically deallocates the intermediate tensors which won't be used again in the remaining operations.Shared memory: When two operations will never be performed at the same time, the result tensors produced by them can share a piece of memory.
How to use
- A CNN example.
class CNN(module.Module):
def __init__(self, optimizer):
super(CNN, self).__init__()
self.conv1 = autograd.Conv2d(1, 20, 5, padding=0)
self.conv2 = autograd.Conv2d(20, 50, 5, padding=0)
self.linear1 = autograd.Linear(4 * 4 * 50, 500)
self.linear2 = autograd.Linear(500, 10)
self.pooling1 = autograd.MaxPool2d(2, 2, padding=0)
self.pooling2 = autograd.MaxPool2d(2, 2, padding=0)
self.optimizer = optimizer
def forward(self, x):
y = self.conv1(x)
y = autograd.relu(y)
y = self.pooling1(y)
y = self.conv2(y)
y = autograd.relu(y)
y = self.pooling2(y)
y = autograd.flatten(y)
y = self.linear1(y)
y = autograd.relu(y)
y = self.linear2(y)
return y
def loss(self, x, ty):
return autograd.softmax_cross_entropy(x, ty)
def optim(self, loss):
self.optimizer.backward_and_update(loss)
# initialization other objects
# ......
model = CNN(sgd)
model.train()
model.on_device(dev)
model.graph(graph, sequential)
# Train
for b in range(num_train_batch):
# Generate the patch data in this iteration
# ......
# Copy the patch data into input tensors
tx.copy_from_numpy(x)
ty.copy_from_numpy(y)
# Train the model
out = model(tx)
loss = model.loss(out, ty)
model.optim(loss)
A Google Colab notebook of this example is available here.
- Some settings:
module.py
training: whether to train the neural network defined in the class or for evaluation.graph_mode: the model class defined by users can be trained using computational graph or not.sequential: execute operations in graph serially or in the order of BFS.
- More examples:
Experiments
Single node
- Experiment settings
- Model
- Using layer: ResNet50 in resnet.py
- Using module: ResNet50 in resnet_module.py
- GPU: NVIDIA RTX 2080Ti
- Model
- Notations
s:secondit: iterationMem:peak memory usage of single GPUThroughout:number of images processed per secondTime:total timeSpeed:iterations per secondReduction:the memory usage reduction rate compared with that using layerSpeedup: speedup ratio compared with dev branch
- Result
Batchsize Cases Mem(MB) Time(s) Speed(it/s) Throughput Reduction Speedup 16 layer 4975 14.1952 14.0893 225.4285 0.00% 1.0000 module:disable graph 4995 14.1264 14.1579 226.5261 -0.40% 1.0049 module:enable graph, bfs 3283 13.7438 14.5520 232.8318 34.01% 1.0328 module:enable graph, serial 3265 13.7420 14.5540 232.8635 34.37% 1.0330 32 layer 10119 13.4587 7.4302 237.7649 0.00% 1.0000 module:enable graph 10109 13.2952 7.5315 240.6875 0.10% 1.0123 module:enable graph, bfs 6839 13.1059 7.6302 244.1648 32.41% 1.0269 module:enable graph, serial 6845 13.0489 7.6635 245.2312 32.35% 1.0314
Multi processes
- Experiment settings
- Model
- using Layer: ResNet50 in resnet_dist.py
- using Module: ResNet50 in resnet_module.py
- GPU: NVIDIA RTX 2080Ti * 2
- MPI: two MPI processes on one node
- Model
- Notations: the same as above
- Result
Batchsize Cases Mem(MB) Time(s) Speed(it/s) Throughput Reduction Speedup 16 layer 5439 17.3323 11.5391 369.2522 0.00% 1.0000 module:disable graph 5427 17.8232 11.2213 359.0831 0.22% 0.9725 module:enable graph, bfs 3389 18.2310 10.9703 351.0504 37.69% 0.9507 module:enable graph, serial 3437 17.0389 11.7378 375.6103 36.81% 1.0172 32 layer 10547 14.8635 6.7279 430.5858 0.00% 1.0000 module:disable graph 10503 14.7746 6.7684 433.1748 0.42% 1.0060 module:enable graph, bfs 6935 14.8553 6.7316 430.8231 34.25% 1.0006 module:enable graph, serial 7027 14.3271 6.9798 446.7074 33.37% 1.0374
Conclusion
- Computational graph does not affect training time and memory usage if the graph is disabled.
- Computational graph can significantly reduce memory usage and training time.
Implementation
Computational graph construction
Buffer the operations: Use the technique of delayed execution to falsely perform operations in the forward propagation and backward propagation once. Buffer all the operations and the tensors read or written by each operation. Take matmul for example.# user calls an api to do matmul on two tensors x = autograd.matmul(inputs, w0) # Python code inside the api singa.Mult(inputs, w)// the backend platform // pass the specific execution function of the operation // and the tensors it will reads and writes during the calculation to the device. C->device()->Exec( [a, A, b, B, CRef](Context *ctx) mutable { GEMV<DType, Lang>(a, A, B, b, &CRef, ctx); }, read_blocks, {C->block()});Build nodes and edges: Build the nodes and edges of the operations passed to the device and add them into the computational graph. Since we just told the scheduler which blocks these operations will read and write and some of the tensors will share the same blocks, the scheduler will split one edge into multiple to ensure that the constructed graph is a directed acyclic graph.Analyze the graph: Calculate dependencies between all the operations to decide the order of execution. The system will only analyze the same graph once. If new operations are added to the graph, the calculation graph will be re-analyzed.Run graph: Execute all the operations in the order we just calculated to update all the parameters. Tensors are well scheduled to allocate and deallocate to save memory. After the analyzing, the operations in the graph can be executed based on the result of analyzing.Module: Provided a module class on the Python side for users to use this feature more conveniently.
Lazy allocation
- When a device needs to create a new block, pass the device to that block only, instead of allocating a piece of memory from the mempool and passing the pointer to that block.
- When a block is accessed for the first time, the device corresponding to the block allocates memory and then access it.
Automatic recycling
- When calculating dependencies between the operations during graph construction, the reference count of tensors can also be calculated.
- When an operation is completed, the schedualer decrease the reference count of tensors that the operation used.
- If a tensor's reference count reaches zero, it means the tensor won't be accessed by latter operations, so we can recycle its memory.
- The program will track the usage of the block. If a block is used on the python side, it will not be recycled, which is convenient for debugging on the python side.
Shared memory
- Once the kernel function of an operation is added into the default cuda stream and the tensors used by the operation can be freed when the calculation is complete, the scheduler will free these tensors' memory immediately and no need to wait for the calculation to complete. Because subsequent operations will not be performed at the same time as the current operation as the platform now used the default stream of CUDA to finish the calculation. So the following tensors can share the same memory with these tensors.
- Use a mempool to manage the GPU memory. Scheduler returns the memory used by tensors to the mempool and the latter tensors will apply for memory from mempool. The mempool will find the most suitable blocks returned by the previous tensors for the latter tensors to share as much memory as possible.
How to add a new operation
For new operations to be included in the computational graph, they should be submitted to the device. Device class on the CPP side will add these operations in the computational graph and the scheduler will schedule them automatically.
Requirements
When submitting operations, there are some requirements.
Need to pass in the function that the operation executes and the data blocks that the operation reads and writes
For the function of the operation: All variables used in lambda expressions need to be captured according to the following rules.
capture by value: If the variable is a local variable or will be immediately released (e.g. intermediate tensors). If not captured by value, these variables will be destroyed after buffering. Buffering is just a way to defer real calculations.capture by reference:If the variable is recorded on the python side or a global variable (e.g. The parameter W and ConvHand in the Conv2d class).mutable: The lambda expression should have mutable tag if a variable captured by value is modified in an expression
Example
- Python side: _Conv2d records x, W, b and handle in the class.
class _Conv2d(Operation):
def __init__(self, handle, odd_padding=(0, 0, 0, 0)):
super(_Conv2d, self).__init__()
self.handle = handle # record handle
self.odd_padding = odd_padding
if self.odd_padding != (0, 0, 0, 0):
self.re_new_handle = True
def forward(self, x, W, b=None):
# other code
# ......
if training:
if self.handle.bias_term:
self.inputs = (x, W, b) # record x, W, b
else:
self.inputs = (x, W)
# other code
# ......
if (type(self.handle) != singa.ConvHandle):
return singa.GpuConvForward(x, W, b, self.handle)
else:
return singa.CpuConvForward(x, W, b, self.handle)
def backward(self, dy):
if (type(self.handle) != singa.ConvHandle):
dx = singa.GpuConvBackwardx(dy, self.inputs[1], self.inputs[0],
self.handle)
dW = singa.GpuConvBackwardW(dy, self.inputs[0], self.inputs[1],
self.handle)
db = singa.GpuConvBackwardb(
dy, self.inputs[2],
self.handle) if self.handle.bias_term else None
else:
dx = singa.CpuConvBackwardx(dy, self.inputs[1], self.inputs[0],
self.handle)
dW = singa.CpuConvBackwardW(dy, self.inputs[0], self.inputs[1],
self.handle)
db = singa.CpuConvBackwardb(
dy, self.inputs[2],
self.handle) if self.handle.bias_term else None
if self.odd_padding != (0, 0, 0, 0):
dx = utils.handle_odd_pad_bwd(dx, self.odd_padding)
if db:
return dx, dW, db
else:
return dx, dW
- C++ side: convolution.cc
Tensor GpuConvBackwardx(const Tensor &dy, const Tensor &W, const Tensor &x,
const CudnnConvHandle &cch) {
CHECK_EQ(dy.device()->lang(), kCuda);
Tensor dx;
dx.ResetLike(x);
dy.device()->Exec(
/*
* dx is a local variable so it's captured by value
* dy is an intermediate tensor and isn't recorded on the python side
* W is an intermediate tensor but it's recorded on the python side
* chh is a variable and it's recorded on the python side
*/
[dx, dy, &W, &cch](Context *ctx) mutable {
Block *wblock = W.block(), *dyblock = dy.block(), *dxblock = dx.block();
float alpha = 1.f, beta = 0.f;
cudnnConvolutionBackwardData(
ctx->cudnn_handle, &alpha, cch.filter_desc, wblock->data(),
cch.y_desc, dyblock->data(), cch.conv_desc, cch.bp_data_alg,
cch.workspace.block()->mutable_data(),
cch.workspace_count * sizeof(float), &beta, cch.x_desc,
dxblock->mutable_data());
},
{dy.block(), W.block()}, {dx.block(), cch.workspace.block()});
/* the lambda expression reads the blocks of tensor dy and w
* and writes the blocks of tensor dx and chh.workspace
*/
return dx;
}
